Note
Go to the end to download the full example code.
MoTIF on simulated data#
This example demonstrates MoTIF [1] on simulated data. In the alphacsc module, we are offering all the alternatives for the users to try. Please cite our paper [2] if you use this implementation.
- [1] Jost, Philippe, et al.
“MoTIF: An efficient algorithm for learning translation invariant dictionaries.” Acoustics, Speech and Signal Processing, 2006. ICASSP 2006 Proceedings. 2006 IEEE International Conference on. Vol. 5. IEEE, 2006.
- [2] Jas, M., Dupr’e La Tour, T., Simsekli, U., & Gramfort, A. (2017).
Learning the Morphology of Brain Signals Using Alpha-Stable Convolutional Sparse Coding. arXiv preprint arXiv:1705.08006.
# Authors: Mainak Jas <mainak.jas@telecom-paristech.fr>
# Tom Dupre La Tour <tom.duprelatour@telecom-paristech.fr>
# Umut Simsekli <umut.simsekli@telecom-paristech.fr>
# Alexandre Gramfort <alexandre.gramfort@telecom-paristech.fr>
#
# License: BSD (3-clause)
Let us first import the modules.
import matplotlib.pyplot as plt
from alphacsc.simulate import simulate_data
from alphacsc.other.motif import learn_atoms
and define the relevant parameters. Note we choose a large n_times to avoid overlapping atoms which MoTIF cannot handle
n_times_atom = 64 # L
n_times = 5000 # T
n_atoms = 2 # K
n_trials = 10 # N
simulate the data.
random_state_simulate = 1
X, ds_true, z_true = simulate_data(n_trials, n_times, n_times_atom,
n_atoms, random_state_simulate,
constant_amplitude=True)
n_times_atom = n_times_atom * 7 # XXX: hack
n_iter = 10
max_shift = 11 # after this, the algorithm breaks
Note, how we use constant_amplitude=True since MoTIF cannot handle atoms of varying amplitudes. Check out our examples on vanilla CSC and alphaCSC to learn how to deal with such cases. Finally, let us estimate the atoms.
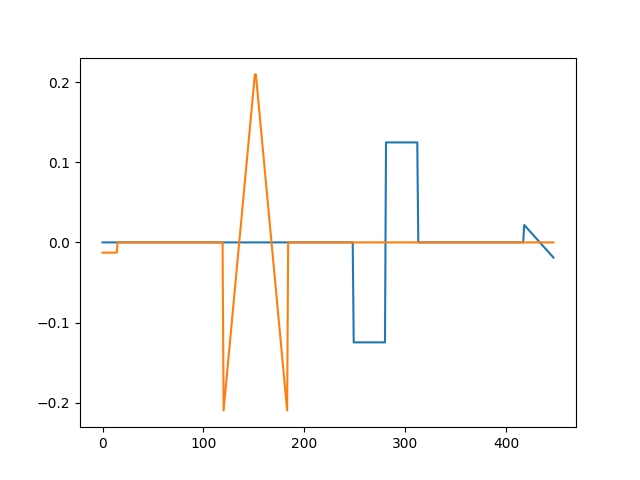
[seed 42] Atom 0 Iteration 0
[seed 42] Atom 0 Iteration 1
[seed 42] Atom 0 Iteration 2
[seed 42] Atom 0 Iteration 3
[seed 42] Atom 0 Iteration 4
[seed 42] Atom 0 Iteration 5
[seed 42] Atom 0 Iteration 6
[seed 42] Atom 0 Iteration 7
[seed 42] Atom 0 Iteration 8
[seed 42] Atom 0 Iteration 9
[seed 42] Atom 1 Iteration 0
[seed 42] Atom 1 Iteration 1
[seed 42] Atom 1 Iteration 2
[seed 42] Atom 1 Iteration 3
[seed 42] Atom 1 Iteration 4
[seed 42] Atom 1 Iteration 5
[seed 42] Atom 1 Iteration 6
[seed 42] Atom 1 Iteration 7
[seed 42] Atom 1 Iteration 8
[seed 42] Atom 1 Iteration 9
[<matplotlib.lines.Line2D object at 0x7f6140e1f520>, <matplotlib.lines.Line2D object at 0x7f6140e1f4f0>]
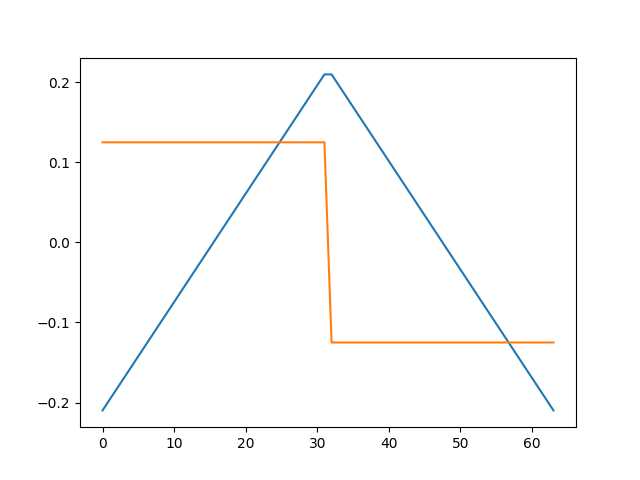
Compare this to the original atoms
plt.figure()
plt.plot(ds_true.T)
plt.show()
Total running time of the script: (0 minutes 2.010 seconds)